Video Article Open Access
Multiplexed Biosensors for Point-of-Care Management of Infectious Diseases
Midori Johnston1,2, H. Ceren Ates1,2, Regina T. Glatz1,2, Hasti Mohsenin3, Rosanne Schmachtenberg3, Nathalie Göppert4, Daniela Huzly4, Gerald A. Urban1,5, Wilfried Weber3, Can Dincer1,2,*
1Department of Microsystems Engineering (IMTEK), University of Freiburg, Freiburg, 79110 Germany
2FIT Freiburg Center for Interactive Materials and Bioinspired Technologies, University of Freiburg, Freiburg, 79110, Germany
3Faculty of Biology and Signalling Research Centers BIOSS and CIBSS, University of Freiburg, Freiburg, 79104, Germany
4Institute of Virology, Medical Center – University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, 79104, Germany
5Freiburg Materials Research Center, University of Freiburg, Freiburg, 79104, Germany
Vid. Proc. Adv. Mater., Volume 3, Article ID 2210351 (2022)
DOI: 10.5185/vpoam.2022.10351
Publication Date (Web): 23 May 2023
Copyright © IAAM
Graphical Abstract
Abstract
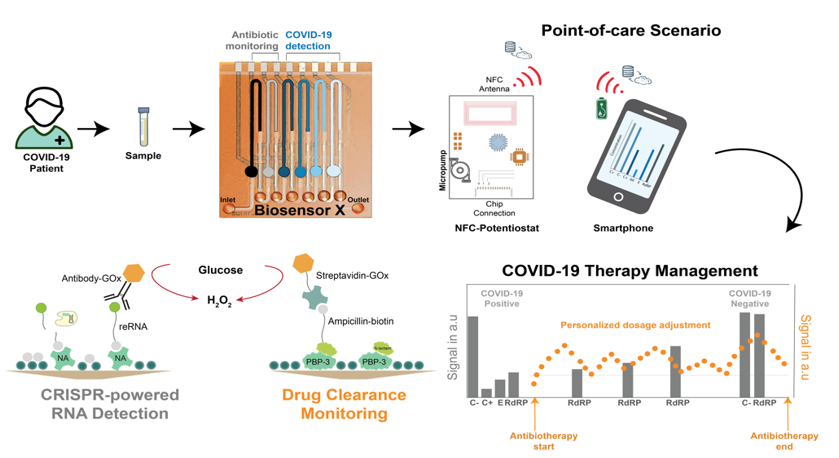
Nucleic acid testing is vital for the diagnostics and treatment monitoring of different diseases in medicine. It primarily involves with the measurement of mutations in DNA sequences, pathogen-specific oligonucleotides or changes in gene expression levels [1]. Over the recent years, especially the (re)emergence outbreak of infectious diseases (such as COVID-19 disease caused by SARS-CoV-2 infections) and finding of nucleic acid-based biomarkers (such as microRNAs) further stimulate the development of novel tools for nucleic acid diagnostics. Herein, CRISPR/Cas technology, broadly applied in gene editing, features a powerful method for the highly sensitive and selective detection of nucleic acids [2]. In this talk, first a short overview of CRISPR-powered biosensing will be given where two different detection modes, binding- and cleavage-based sensing, will be discussed2. Subsequently, an electrochemical microfluidic multiplexed biosensor (BiosensorX) platform for point-of-care management of COVID-19 will be introduced (see Graphical Abstract) [3,4,5]. The technology developed is capable of gauging 4, 6, or 8 (different) analytes or specimen simultaneously via a sequential chip design, including multiple immobilization areas, where the assay reagents are simply adsorbed, followed by their individual electrochemical measurement cells, where the signal readout takes place by means of amperometry, within a single microfluidic channel. In this study, different (RdRp and E) genes of SARS-CoV-2 (Omicron-variant) in clinical samples is simultaneously screened and identified by employing the enzyme LbuCas13a. By eliminating the need for a reverse transcription and nucleic acid amplification of the viral RNAs (which is necessary for PCR and other detection methods), an LOD of 2,000 copies per µl (4.06 fM) for the E gene and 7,520 copies per µl (15.04 fM) for RdRP is achieved within a sample-to-result time of about 30 minutes. Moreover, in order to demonstrate the feasibility of combining assays based on different classes of biomolecules (here, nucleic acids along with drugs), antibody-free ß-lactam antibiotic detection using a penicillin-binding protein-based bioassay is performed on the same biosensor device. Without the need for any target amplification, BiosensorX platform offers an affordable, easily scalable and multiplexed approach for point-of-care testing of nucleic acids via CRISPR-based bioassays and other biomolecule classes (such as drugs) at the same time.
Keywords
CRISPR diagnostics; COVID-19; antibiotics; multiplexed microfluidics; electrochemical biosensors.
Acknowledgement
The authors would like to thank the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) and Bundesministerium für Bildung und Forschung (BMBF, Federal Ministry of Education and Research) for partially funding this work under grant numbers 404478562, 421356369, 390939984 (EXC 2189 – CIBSS), and 13GW0493 (MERGE).
References
- E. Morales-Narváez, C. Dincer, Biosensors and Bioelectronics, 2020, 163, 112274.
- Q.A Phan, L.B. Truong, D. Medina-Cruz, C. Dincer, E. Mostafavi, Biosensors and Bioelectronics, 2022, 197, 113732.
- R. Bruch, M. Johnston, A. Kling, T. Mattmüller, J. Baaske, S. Partel, S. Madlener, W. Weber, G.A. Urban, C. Dincer, Biosensors and Bioelectronics, 2021, 177, 112887.
- R.T. Glatz, H.C. Ates, H. Mohsenin, W. Weber, C. Dincer, Analytical and Bioanalytical Chemistry, 2022, https://doi.org/10.1007/s00216-022-04210-4.
- M. Johnston, H.C. Ates, R.T. Glatz, H. Mohsenin, R. Schmachtenberg, N. Göppert, D. Huzly, G.A. Urban, W. Weber, C. Dincer, MedRxiv, 2022, https://doi.org/10.1101/2022.03.30.22271928.
Biography
Can Dincer is currently junior research group leader at FIT Freiburg Center for Interactive Materials and Bioinspired Technologies and at the Department of Microsystems Engineering (IMTEK) of the University of Freiburg. The main research interest of his group "Disposable Microsystems" is the development of bioanalytical materials/sensors/ microsystems and their combination with data science and artificial intelligence for various applications including point-of-care diagnostics, wearables, food and environmental analysis. Having completed his studies in microsystems engineering, Dr. Dincer received in 2016 his Ph.D. degree with summa cum laude from the University of Freiburg. Between June 2017 - June 2019, he also worked as a visiting researcher at the Department of Bioengineering at the Imperial College London. Since September 2019, he is also an Associate Editor of the journal “Biosensors and Bioelectronics” (Elsevier). In June 2022, he has joined to the International Advisory Board of the new journal “Advanced Sensor Research” (Wiley).
Video Proceedings of Advanced Materials

Upcoming Congress



